How can I evaluate the hydrophobicity of a protein with PyMol?
Open Pymol and load up your protein of choice. Download color_h.py which is a script from the University of Osaka that colours the residues according to Eisenberg's scale of hydrophobicity. Load this into Pymol by File->Run->PATH/TO/color_h.py. Then in Pymol run the following commands:
show surface
color_h obj01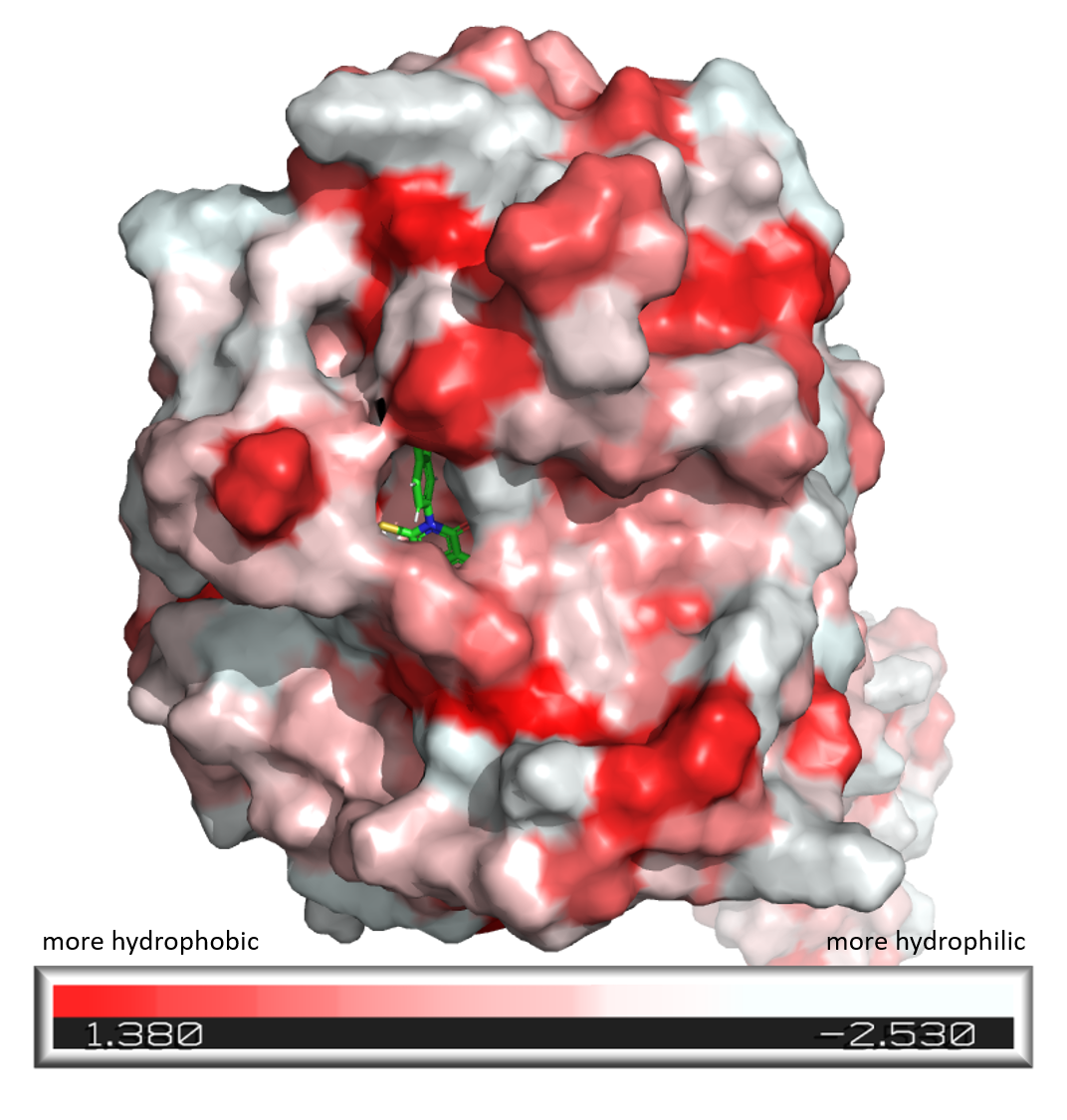
from pymol import cmd
# Apply color per residue type within the selection
# usage: color_h my_obj01
def color_h(selection='all'):
s = str(selection)
print(f"Coloring selection: {s}")
# Define hydrophobicity colors
color_map = {
'ile': [0.996, 0.062, 0.062],
'phe': [0.996, 0.109, 0.109],
'val': [0.992, 0.156, 0.156],
'leu': [0.992, 0.207, 0.207],
'trp': [0.992, 0.254, 0.254],
'met': [0.988, 0.301, 0.301],
'ala': [0.988, 0.348, 0.348],
'gly': [0.984, 0.394, 0.394],
'cys': [0.984, 0.445, 0.445],
'tyr': [0.984, 0.492, 0.492],
'pro': [0.980, 0.539, 0.539],
'thr': [0.980, 0.586, 0.586],
'ser': [0.980, 0.637, 0.637],
'his': [0.977, 0.684, 0.684],
'glu': [0.977, 0.730, 0.730],
'asn': [0.973, 0.777, 0.777],
'gln': [0.973, 0.824, 0.824],
'asp': [0.973, 0.875, 0.875],
'lys': [0.899, 0.922, 0.922],
'arg': [0.899, 0.969, 0.969],
}
for resn, rgb in color_map.items():
color_name = f'color_{resn}'
cmd.set_color(color_name, rgb)
cmd.color(color_name, f"({s}) and resn {resn}")
cmd.extend('color_h', color_h)# color_h
# -------
# PyMOL command to color protein molecules according to the Eisenberg hydrophobicity scale
#
# Source: http://us.expasy.org/tools/pscale/Hphob.Eisenberg.html
# Amino acid scale: Normalized consensus hydrophobicity scale
# Author(s): Eisenberg D., Schwarz E., Komarony M., Wall R.
# Reference: J. Mol. Biol. 179:125-142 (1984)
# https://protein-sol.manchester.ac.uk/
# https://www.nature.com/articles/s41598-018-36950-8
# Amino acid scale values:
#
# Ile: 1.38 [0.996,0.062,0.062]
# Phe: 1.19 [0.996,0.109,0.109]
# Val: 1.08 [0.992,0.156,0.156]
# Leu: 1.06 [0.992,0.207,0.207]
# Trp: 0.81 [0.992,0.254,0.254]
# Met: 0.64 [0.988,0.301,0.301]
# Ala: 0.62 [0.988,0.348,0.348]
# Gly: 0.48 [0.984,0.394,0.394]
# Cys: 0.29 [0.984,0.445,0.445]
# Tyr: 0.26 [0.984,0.492,0.492]
# Pro: 0.12 [0.980,0.539,0.539]
# Thr: -0.05 [0.980,0.586,0.586]
# Ser: -0.18 [0.980,0.637,0.637]
# His: -0.4 [0.977,0.684,0.684]
# Glu: -0.74 [0.977,0.730,0.730]
# Asn: -0.78 [0.973,0.777,0.777]
# Gln: -0.85 [0.973,0.824,0.824]
# Asp: -0.9 [0.973,0.875,0.875]
# Lys: -1.5 [0.899,0.922,0.922]
# Arg: -2.53 [0.899,0.969,0.969]
#
# load https://shaker.umh.es/computing/hydrophobic.pdb
# extract ligand, organic
# show surface
# File->Run->PATH/TO/color_h.py
# color_h
# ramp_new prox, ligand, [1.38, 1.19, 1.08, 1.06, 0.81, 0.64, 0.62, 0.48, 0.29, 0.26, 0.12, -0.05, -0.18, -0.4, -0.74, -0.78, -0.85, -0.9, -1.5, -2.53], [[0.996,0.062,0.062], [0.996,0.109,0.109], [0.992,0.156,0.156], [0.992,0.207,0.207], [0.992,0.254,0.254], [0.988,0.301,0.301], [0.988,0.348,0.348], [0.984,0.394,0.394], [0.984,0.445,0.445], [0.984,0.492,0.492], [0.980,0.539,0.539], [0.980,0.586,0.586], [0.980,0.637,0.637], [0.977,0.684,0.684], [0.977,0.730,0.730], [0.973,0.777,0.777], [0.973,0.824,0.824], [0.973,0.875,0.875], [0.899,0.922,0.922], [0.899,0.969,0.969]]
#
# Usage:
# color_h (selection)
#
from pymol import cmd
def color_h(selection='all'):
s = str(selection)
print s
cmd.set_color('color_ile',[0.996,0.062,0.062])
cmd.set_color('color_phe',[0.996,0.109,0.109])
cmd.set_color('color_val',[0.992,0.156,0.156])
cmd.set_color('color_leu',[0.992,0.207,0.207])
cmd.set_color('color_trp',[0.992,0.254,0.254])
cmd.set_color('color_met',[0.988,0.301,0.301])
cmd.set_color('color_ala',[0.988,0.348,0.348])
cmd.set_color('color_gly',[0.984,0.394,0.394])
cmd.set_color('color_cys',[0.984,0.445,0.445])
cmd.set_color('color_tyr',[0.984,0.492,0.492])
cmd.set_color('color_pro',[0.980,0.539,0.539])
cmd.set_color('color_thr',[0.980,0.586,0.586])
cmd.set_color('color_ser',[0.980,0.637,0.637])
cmd.set_color('color_his',[0.977,0.684,0.684])
cmd.set_color('color_glu',[0.977,0.730,0.730])
cmd.set_color('color_asn',[0.973,0.777,0.777])
cmd.set_color('color_gln',[0.973,0.824,0.824])
cmd.set_color('color_asp',[0.973,0.875,0.875])
cmd.set_color('color_lys',[0.899,0.922,0.922])
cmd.set_color('color_arg',[0.899,0.969,0.969])
cmd.color("color_ile","("+s+" and resn ile)")
cmd.color("color_phe","("+s+" and resn phe)")
cmd.color("color_val","("+s+" and resn val)")
cmd.color("color_leu","("+s+" and resn leu)")
cmd.color("color_trp","("+s+" and resn trp)")
cmd.color("color_met","("+s+" and resn met)")
cmd.color("color_ala","("+s+" and resn ala)")
cmd.color("color_gly","("+s+" and resn gly)")
cmd.color("color_cys","("+s+" and resn cys)")
cmd.color("color_tyr","("+s+" and resn tyr)")
cmd.color("color_pro","("+s+" and resn pro)")
cmd.color("color_thr","("+s+" and resn thr)")
cmd.color("color_ser","("+s+" and resn ser)")
cmd.color("color_his","("+s+" and resn his)")
cmd.color("color_glu","("+s+" and resn glu)")
cmd.color("color_asn","("+s+" and resn asn)")
cmd.color("color_gln","("+s+" and resn gln)")
cmd.color("color_asp","("+s+" and resn asp)")
cmd.color("color_lys","("+s+" and resn lys)")
cmd.color("color_arg","("+s+" and resn arg)")
cmd.extend('color_h',color_h)
def color_h2(selection='all'):
s = str(selection)
print s
cmd.set_color("color_ile2",[0.938,1,0.938])
cmd.set_color("color_phe2",[0.891,1,0.891])
cmd.set_color("color_val2",[0.844,1,0.844])
cmd.set_color("color_leu2",[0.793,1,0.793])
cmd.set_color("color_trp2",[0.746,1,0.746])
cmd.set_color("color_met2",[0.699,1,0.699])
cmd.set_color("color_ala2",[0.652,1,0.652])
cmd.set_color("color_gly2",[0.606,1,0.606])
cmd.set_color("color_cys2",[0.555,1,0.555])
cmd.set_color("color_tyr2",[0.508,1,0.508])
cmd.set_color("color_pro2",[0.461,1,0.461])
cmd.set_color("color_thr2",[0.414,1,0.414])
cmd.set_color("color_ser2",[0.363,1,0.363])
cmd.set_color("color_his2",[0.316,1,0.316])
cmd.set_color("color_glu2",[0.27,1,0.27])
cmd.set_color("color_asn2",[0.223,1,0.223])
cmd.set_color("color_gln2",[0.176,1,0.176])
cmd.set_color("color_asp2",[0.125,1,0.125])
cmd.set_color("color_lys2",[0.078,1,0.078])
cmd.set_color("color_arg2",[0.031,1,0.031])
cmd.color("color_ile2","("+s+" and resn ile)")
cmd.color("color_phe2","("+s+" and resn phe)")
cmd.color("color_val2","("+s+" and resn val)")
cmd.color("color_leu2","("+s+" and resn leu)")
cmd.color("color_trp2","("+s+" and resn trp)")
cmd.color("color_met2","("+s+" and resn met)")
cmd.color("color_ala2","("+s+" and resn ala)")
cmd.color("color_gly2","("+s+" and resn gly)")
cmd.color("color_cys2","("+s+" and resn cys)")
cmd.color("color_tyr2","("+s+" and resn tyr)")
cmd.color("color_pro2","("+s+" and resn pro)")
cmd.color("color_thr2","("+s+" and resn thr)")
cmd.color("color_ser2","("+s+" and resn ser)")
cmd.color("color_his2","("+s+" and resn his)")
cmd.color("color_glu2","("+s+" and resn glu)")
cmd.color("color_asn2","("+s+" and resn asn)")
cmd.color("color_gln2","("+s+" and resn gln)")
cmd.color("color_asp2","("+s+" and resn asp)")
cmd.color("color_lys2","("+s+" and resn lys)")
cmd.color("color_arg2","("+s+" and resn arg)")
cmd.extend('color_h2',color_h2)