How can I generate a nice picture by pymol?
This is the online supplemental wiki for "An Introduction to Biomolecular Graphics" PLoS Comput Biol (2010) 6(8): e1000918. doi:10.1371/journal.pcbi.1000918.
Cool PyMOL-generated Images and their Scripts.
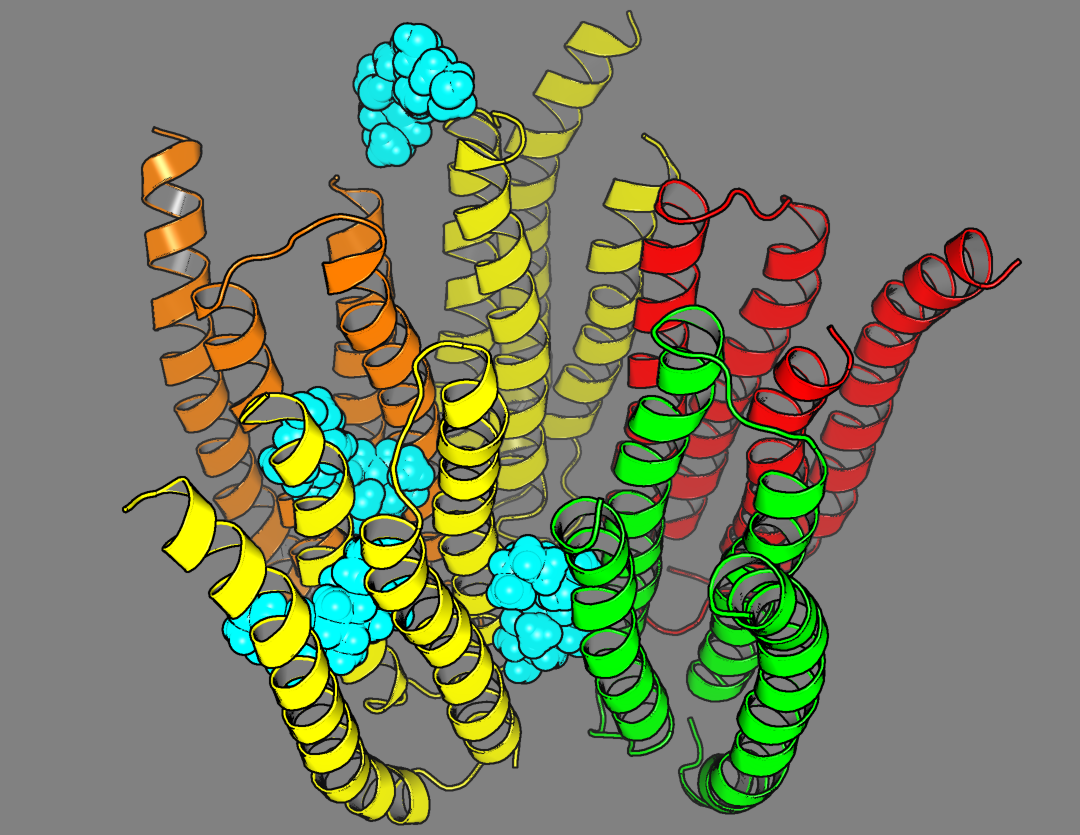 |
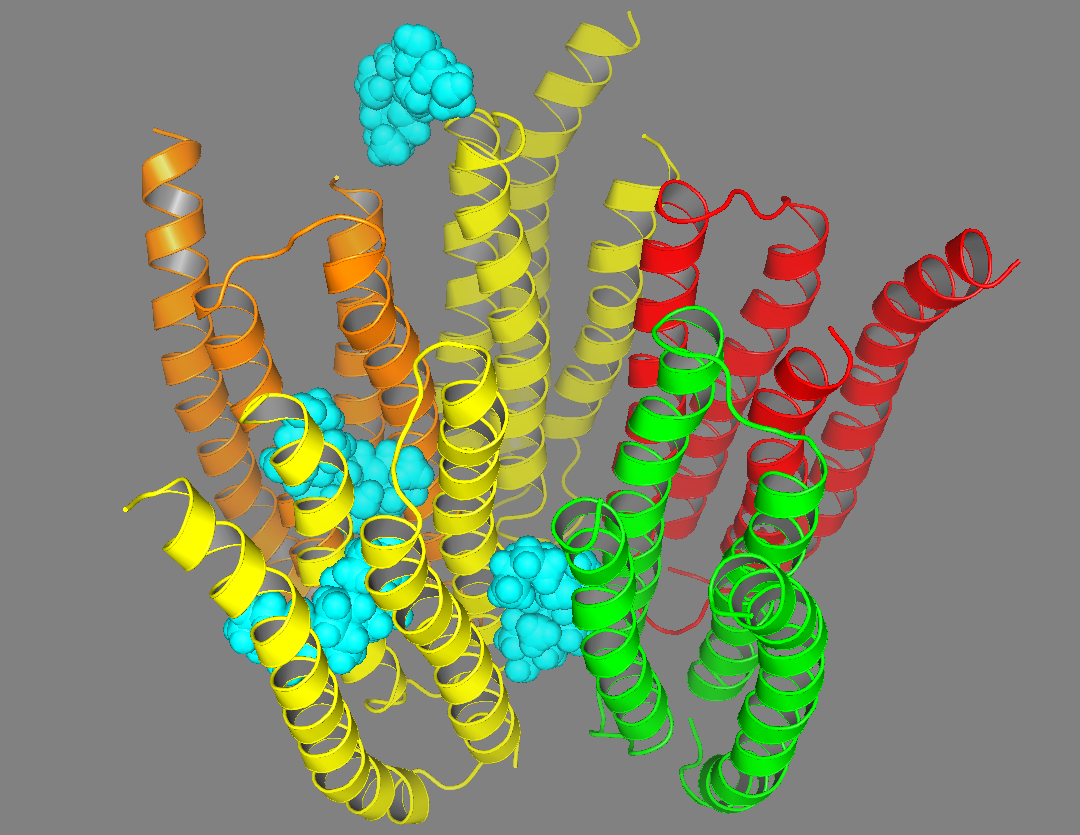 |
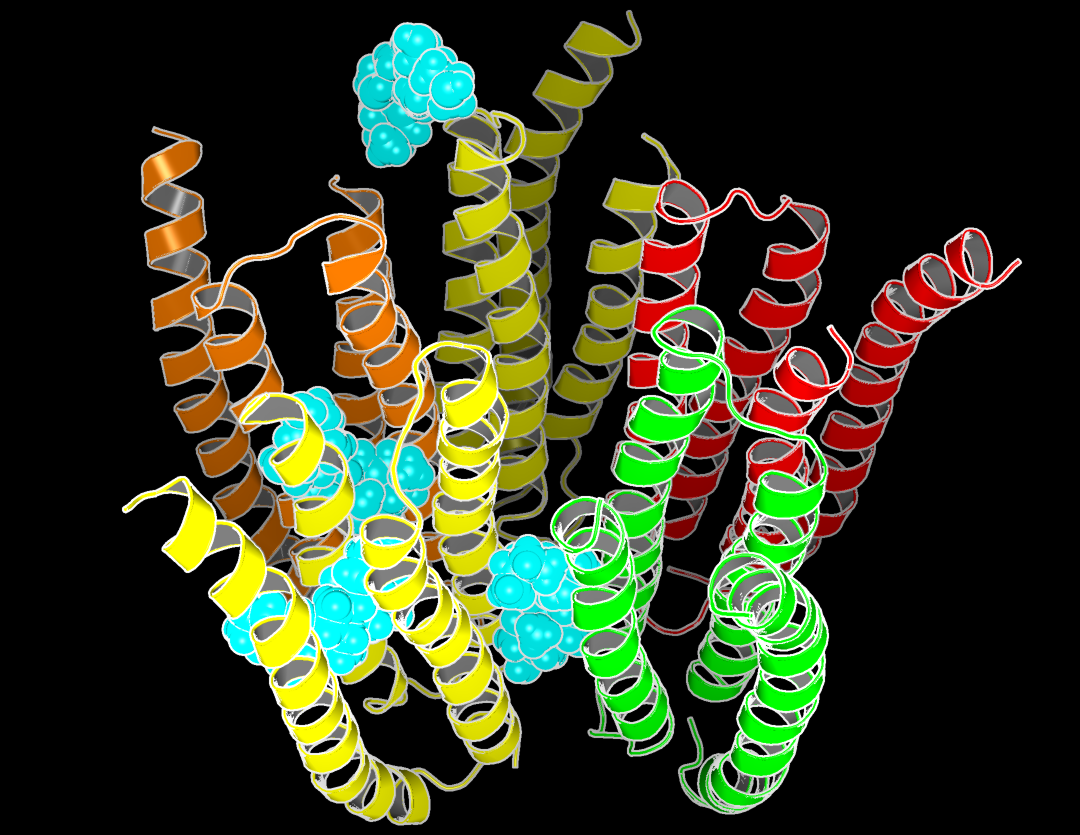 |
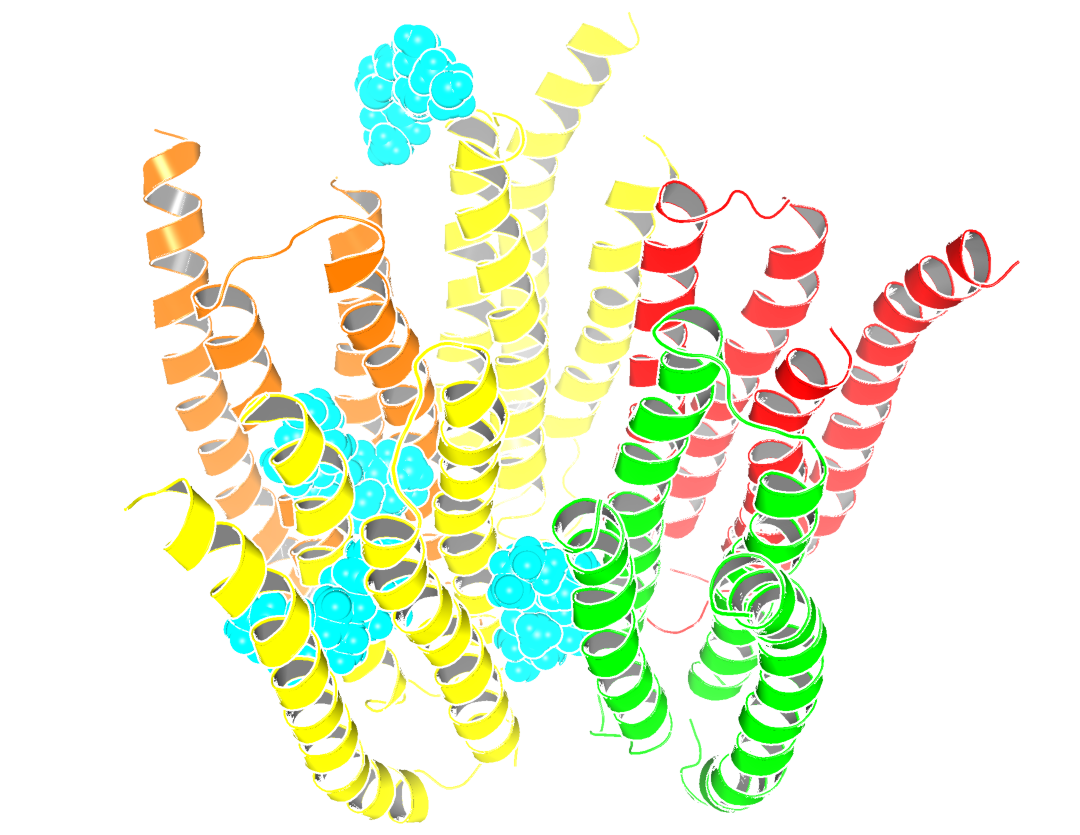 |
# antialias =1 smooths jagged edges, 0 turns it off
set antialias = 1
# Larger values of ambient make the image brighter
set ambient=0.3
# Larger values of direct eliminates shadows
set direct=1.0
set ribbon_radius =0.2
# cartoon_highlight_color will give a separate color to edges of secondary structural elements.
set cartoon_highlight_color =grey50
# ray_trace_mode=1 makes a black outline around the secondary structural elements
set ray_trace_mode=1
# stick_radius -adjust thickness of atomic bonds
set stick_radius = 0.2
# mesh_radius -to adjust thickness of electron density contours
set mesh_radius = 0.02
# bg_color --set the background color
bg_color white
# load pdb file and give it an object name
load https://shaker.umh.es/computing/4aq9-ligands.pdb
# hide nonbonded atoms (i.e. waters)
hide nonbonded
# show cartoon ribbons
show cartoon
# Hide the default line representation of atomic bonds
hide lines
# Use standard helix, strand, and loop representations
# other possibilities: cartoon loop, cartoon rect,
# cartoon oval, and cartoon tube
set cartoon_tube_radius, 0.2
# Make fancy helices with ridge on the edges like
# molscript does
# 1 is on. 0 is off.
set cartoon_fancy_helices=1
set cartoon_cylindrical_helices=0
# Make the strands flat=1 or pass through CA positions=0
# Set to 0 when showing side chains from a strand
set cartoon_flat_sheets = 1.0
# Draw the loops smooth=1 or pass through CA positions=0
# Set to 0 when showing side chains from a loop
set cartoon_smooth_loops = 0
# Example loading a protein and its electron density map
# fetch 1cll
load https://shaker.umh.es/computing/4aq9-ligands.pdb
# set object's color to colorName
set label_color, blue, coco
# set the label size to 10pt
set label_size, 10
# set object's color to colorName
set label_color, colorName, object
# label_position sets the offset of the label relative to the atomic position.
# Offsets the labels 3 Å in X, 2 in Y and 1 in Z
set label_position,(3,2,1)