How to align the sequences of several proteins shown in a PyMol scene?
I have two structures of related organisms. I want to align them in 3D with PyMol and be able to view their sequences aligned as well (corresponding to structure alignment).
set seq_view, on
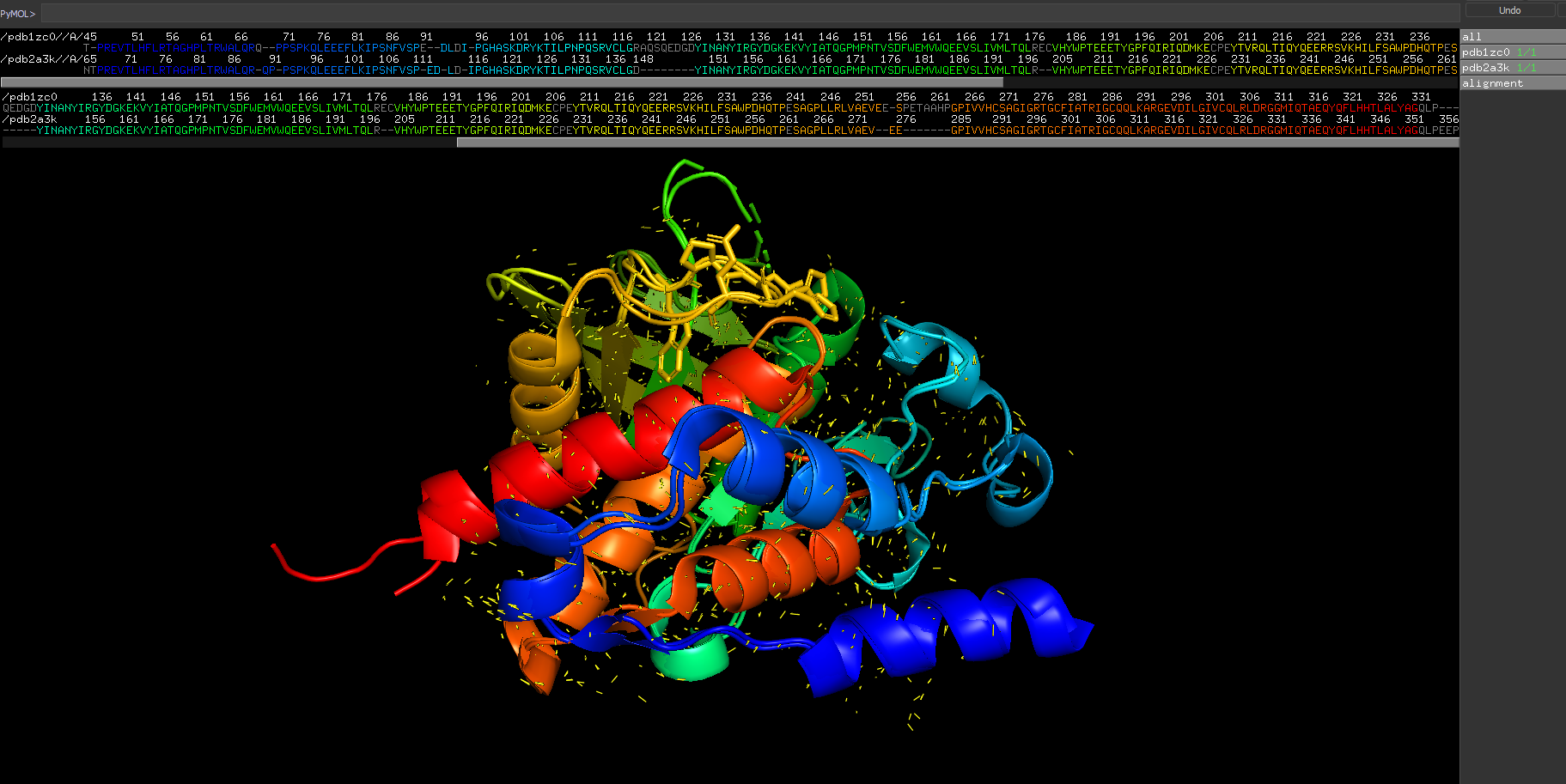
super obj1, obj2, object=alignmentThis shows CGO lines between the paired atoms on the structures, and also aligns the sequences (where the grey letters are non-matching). Many thanks to Jason Vertrees.
In the following PyMol scene we have aligned 12 tyrosine-protein phosphatase non-receptor type 7 structures. However, the amino acid sequences are not shown aligned.
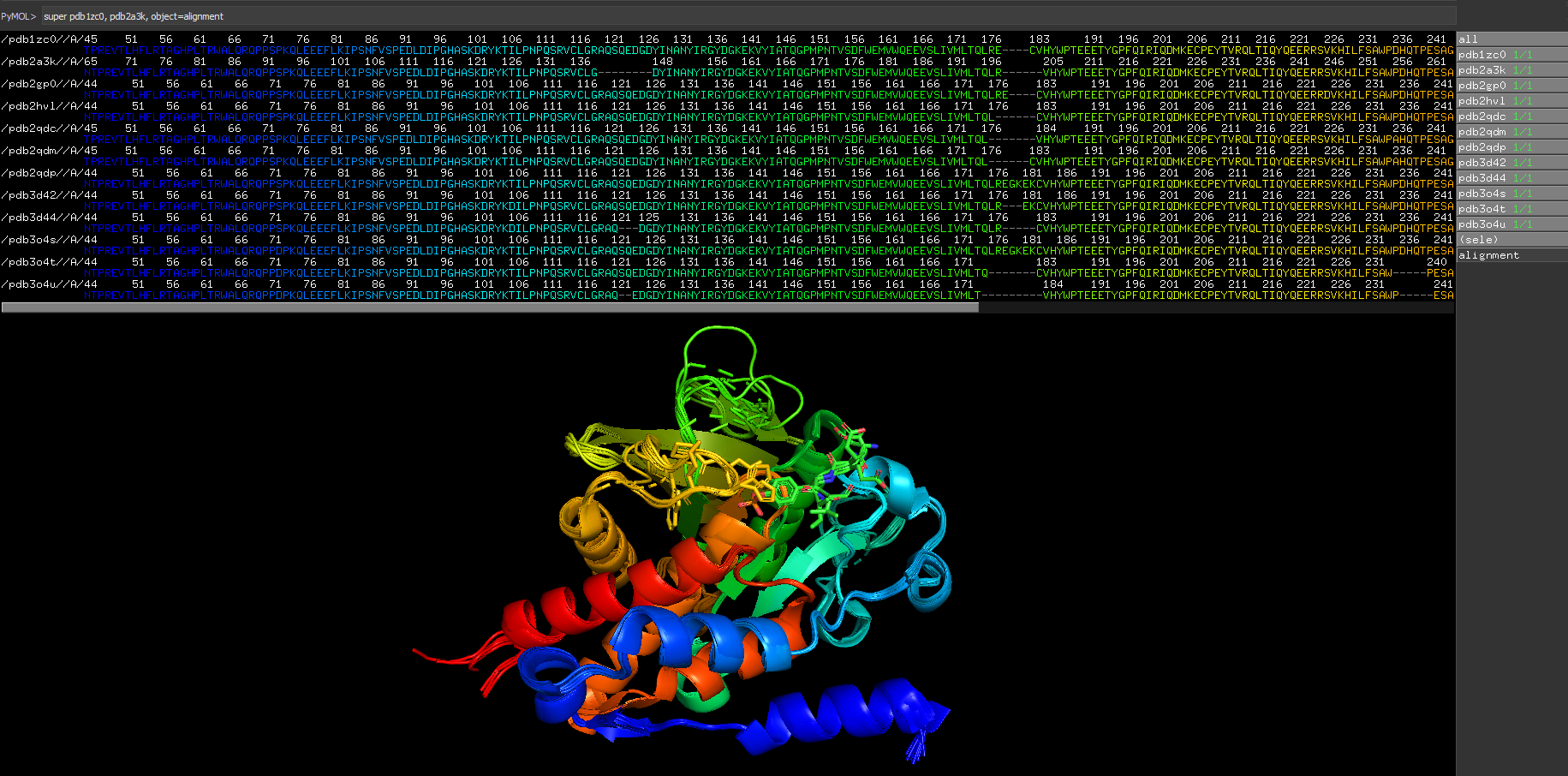
super obj1, obj2, object=alignment