PyMol tutorials by James J. Ross
# color_h
# -------
# PyMOL command to color protein molecules according to the Eisenberg hydrophobicity scale
# Source: http://us.expasy.org/tools/pscale/Hphob.Eisenberg.html
# Amino acid scale: Normalized consensus hydrophobicity scale
# Author(s): Eisenberg D., Schwarz E., Komarony M., Wall R.
# Reference: J. Mol. Biol. 179:125-142 (1984)
# Amino acid scale values:
# Ala: 0.620
# Arg: -2.530
# Asn: -0.780
# Asp: -0.900
# Cys: 0.290
# Gln: -0.850
# Glu: -0.740
# Gly: 0.480
# His: -0.400
# Ile: 1.380
# Leu: 1.060
# Lys: -1.500
# Met: 0.640
# Phe: 1.190
# Pro: 0.120
# Ser: -0.180
# Thr: -0.050
# Trp: 0.810
# Tyr: 0.260
# Val: 1.080
# Usage:
# color_h (selection)
from pymol import cmd
def color_h(selection='all'):
s = str(selection)
print(s)
cmd.set_color('color_ile',[0.996,0.062,0.062])
cmd.set_color('color_phe',[0.996,0.109,0.109])
cmd.set_color('color_val',[0.992,0.156,0.156])
cmd.set_color('color_leu',[0.992,0.207,0.207])
cmd.set_color('color_trp',[0.992,0.254,0.254])
cmd.set_color('color_met',[0.988,0.301,0.301])
cmd.set_color('color_ala',[0.988,0.348,0.348])
cmd.set_color('color_gly',[0.984,0.394,0.394])
cmd.set_color('color_cys',[0.984,0.445,0.445])
cmd.set_color('color_tyr',[0.984,0.492,0.492])
cmd.set_color('color_pro',[0.980,0.539,0.539])
cmd.set_color('color_thr',[0.980,0.586,0.586])
cmd.set_color('color_ser',[0.980,0.637,0.637])
cmd.set_color('color_his',[0.977,0.684,0.684])
cmd.set_color('color_glu',[0.977,0.730,0.730])
cmd.set_color('color_asn',[0.973,0.777,0.777])
cmd.set_color('color_gln',[0.973,0.824,0.824])
cmd.set_color('color_asp',[0.973,0.875,0.875])
cmd.set_color('color_lys',[0.899,0.922,0.922])
cmd.set_color('color_arg',[0.899,0.969,0.969])
cmd.color("color_ile","("+s+" and resn ile)")
cmd.color("color_phe","("+s+" and resn phe)")
cmd.color("color_val","("+s+" and resn val)")
cmd.color("color_leu","("+s+" and resn leu)")
cmd.color("color_trp","("+s+" and resn trp)")
cmd.color("color_met","("+s+" and resn met)")
cmd.color("color_ala","("+s+" and resn ala)")
cmd.color("color_gly","("+s+" and resn gly)")
cmd.color("color_cys","("+s+" and resn cys)")
cmd.color("color_tyr","("+s+" and resn tyr)")
cmd.color("color_pro","("+s+" and resn pro)")
cmd.color("color_thr","("+s+" and resn thr)")
cmd.color("color_ser","("+s+" and resn ser)")
cmd.color("color_his","("+s+" and resn his)")
cmd.color("color_glu","("+s+" and resn glu)")
cmd.color("color_asn","("+s+" and resn asn)")
cmd.color("color_gln","("+s+" and resn gln)")
cmd.color("color_asp","("+s+" and resn asp)")
cmd.color("color_lys","("+s+" and resn lys)")
cmd.color("color_arg","("+s+" and resn arg)")
cmd.extend('color_h',color_h)
def color_h2(selection='all'):
s = str(selection)
print(s)
cmd.set_color("color_ile2",[0.938,1,0.938])
cmd.set_color("color_phe2",[0.891,1,0.891])
cmd.set_color("color_val2",[0.844,1,0.844])
cmd.set_color("color_leu2",[0.793,1,0.793])
cmd.set_color("color_trp2",[0.746,1,0.746])
cmd.set_color("color_met2",[0.699,1,0.699])
cmd.set_color("color_ala2",[0.652,1,0.652])
cmd.set_color("color_gly2",[0.606,1,0.606])
cmd.set_color("color_cys2",[0.555,1,0.555])
cmd.set_color("color_tyr2",[0.508,1,0.508])
cmd.set_color("color_pro2",[0.461,1,0.461])
cmd.set_color("color_thr2",[0.414,1,0.414])
cmd.set_color("color_ser2",[0.363,1,0.363])
cmd.set_color("color_his2",[0.316,1,0.316])
cmd.set_color("color_glu2",[0.27,1,0.27])
cmd.set_color("color_asn2",[0.223,1,0.223])
cmd.set_color("color_gln2",[0.176,1,0.176])
cmd.set_color("color_asp2",[0.125,1,0.125])
cmd.set_color("color_lys2",[0.078,1,0.078])
cmd.set_color("color_arg2",[0.031,1,0.031])
cmd.color("color_ile2","("+s+" and resn ile)")
cmd.color("color_phe2","("+s+" and resn phe)")
cmd.color("color_val2","("+s+" and resn val)")
cmd.color("color_leu2","("+s+" and resn leu)")
cmd.color("color_trp2","("+s+" and resn trp)")
cmd.color("color_met2","("+s+" and resn met)")
cmd.color("color_ala2","("+s+" and resn ala)")
cmd.color("color_gly2","("+s+" and resn gly)")
cmd.color("color_cys2","("+s+" and resn cys)")
cmd.color("color_tyr2","("+s+" and resn tyr)")
cmd.color("color_pro2","("+s+" and resn pro)")
cmd.color("color_thr2","("+s+" and resn thr)")
cmd.color("color_ser2","("+s+" and resn ser)")
cmd.color("color_his2","("+s+" and resn his)")
cmd.color("color_glu2","("+s+" and resn glu)")
cmd.color("color_asn2","("+s+" and resn asn)")
cmd.color("color_gln2","("+s+" and resn gln)")
cmd.color("color_asp2","("+s+" and resn asp)")
cmd.color("color_lys2","("+s+" and resn lys)")
cmd.color("color_arg2","("+s+" and resn arg)")
cmd.extend('color_h2',color_h2) |
|
 |
 |
niceify2.pml |
![]() PyMOL-1-GUI.pdf
PyMOL-1-GUI.pdf
![]() Tutorial-6-FAQ.txt
Tutorial-6-FAQ.txt
Labels location
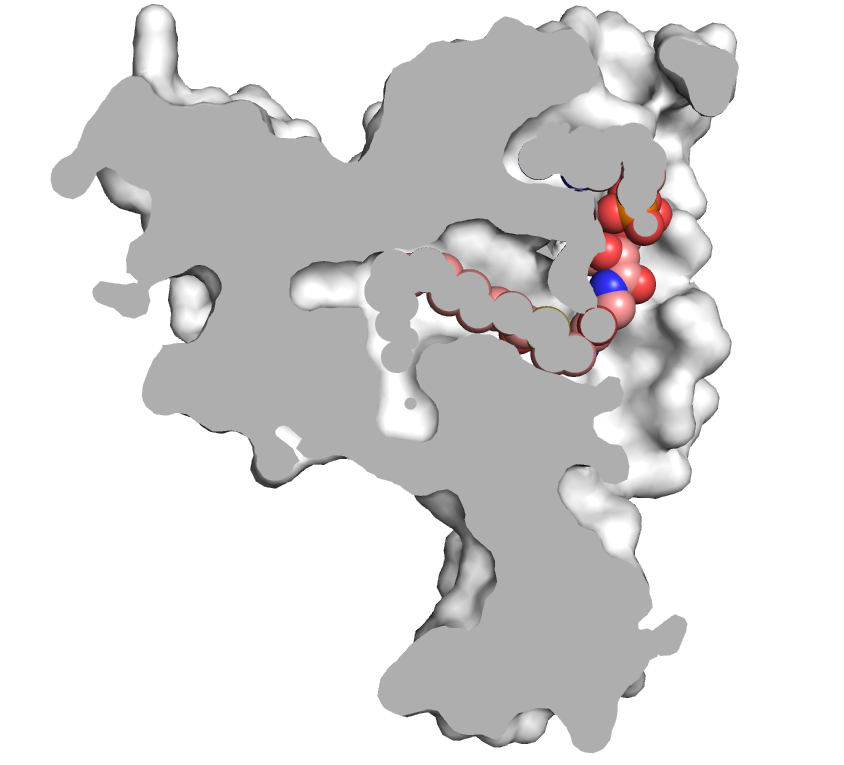
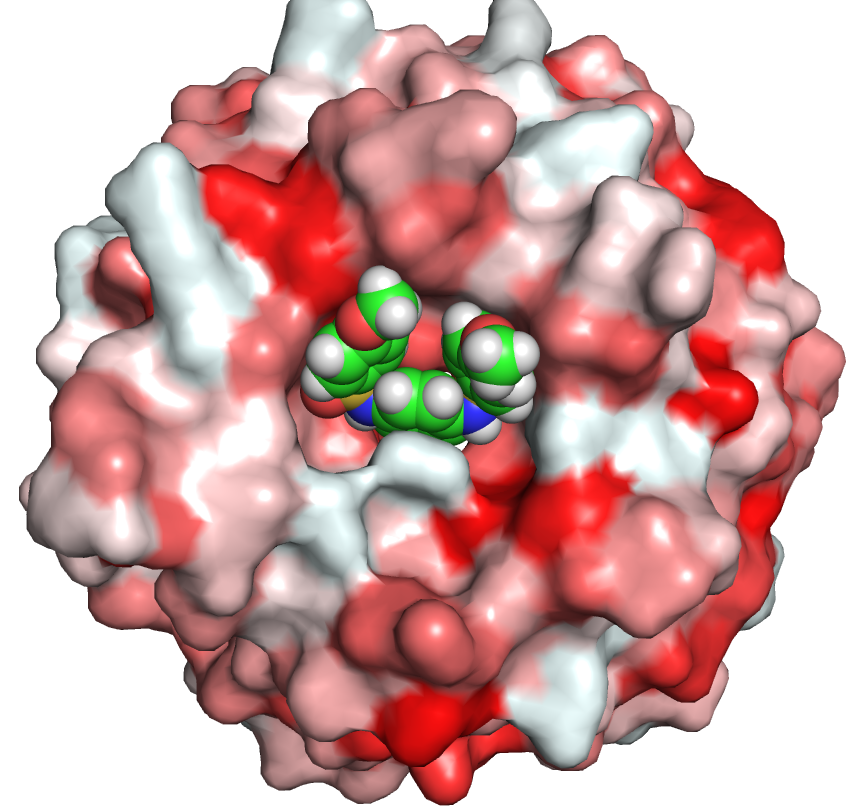
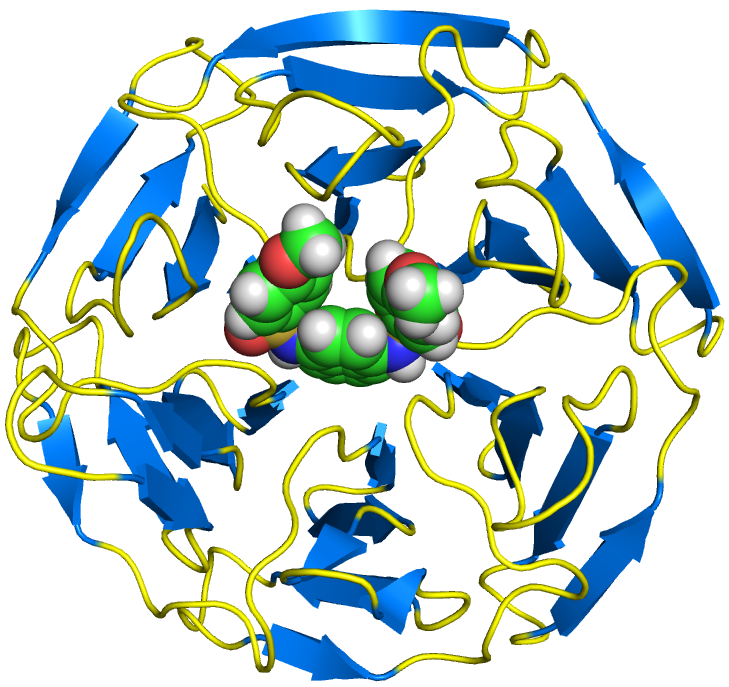
delta9-desaturase2.pml
reinitialize
fetch 4zyo, async=0
# set background
bg_color white
# make selections and objects
extract lig, organic
select bb, bb.
select sc, sc. + n. ca + PRO/N
select ligand, lig
# clean and show
remove hydrogen + solvent
hide all
as cartoon
show sticks, sc
show sticks, lig
util.cbc
color salmon, lig
util.cnc
orient bb
select none
set_view (\
0.359103978, -0.708514035, -0.607498288,\
0.471494555, 0.699466527, -0.537065685,\
0.805441797, -0.093569458, 0.585241377,\
-0.000046019, -0.000023372, -182.910034180,\
21.997421265, 64.741020203, 40.453483582,\
146.089828491, 219.724777222, -20.000000000 )
set hash_max, 400
# must disable depth cue and shadows
set depth_cue, 0
set ray_shadows, 0
set ray_trace_mode, 0
select tirar, /lig/E/A/LMT
remove tirar
delete tirar
set hash_max, 400
# must disable depth cue and shadows
set depth_cue, 0
set ray_shadows, 0
set ray_trace_mode, 0
# this controls the z depth of the slice plane
# (sets it halfway between the clipping planes)
fraction = 0.42
view = cmd.get_view()
near_dist = fraction*(view[16]-view[14])
far_dist = (view[16]-view[15]) - near_dist
cmd.clip("near", -near_dist)
# render opaque background image
as surface
set ray_interior_color, grey90
set opaque_background
set surface_color, white
ray
set_view (\
0.097542562, -0.798652232, -0.593834102,\
-0.104675643, 0.585134983, -0.804151118,\
0.989709318, 0.140598804, -0.026524151,\
-0.000109777, -0.000140121, -159.775024414,\
22.167934418, 64.722122192, 37.309017181,\
161.295089722, 189.170837402, -20.000000000 )
select tirar, /lig/D/A/ST9
show spheres, tirar
png dpi600-protein, dpi=600, ray=1
hide surface, tirar
ray