Aplicaremos el macro de YASARA dock_runscreening.mcr cuyas primeras líneas ves justo debajo.
#MacroTarget '/home/myname/projects/docking/1sdf'
ph=7.4
# Docking method, either VINA (CPU only) or AutoDockLGA (runs on the GPU if enabled at Options > Processors > Set compute GPU)
method='AutoDockLGA'
# Number of docking runs per ligand (maximally 999, 0 lets YASARA choose based on the number of CPUs)
runs=999
# Number of best poses per ligand saved and reported
bestposes=1
# Set to 1 to keep the ligand completely rigid (alternatively you can provide
# the ligand as a *.yob file and fix certain dihedral angles only).
rigid=0
# A selection of receptor residues whose side-chains should be kept flexible, e.g.
flexres=''
# Sort results either by ligand number ('ligandnum'), binding energy ('bindnrg') or by
# efficiency ('effi'), i.e. binding energy per heavy atom, which addresses the bias towards
# larger ligands in screening.
sortby='ligandnum'
# Force field used for charge assignment (AutoDock only, not VINA) and to determine which ligand bonds can rotate.
# Do not change to an in vacuo force field (NOVA), all other force fields should work too (not tested).
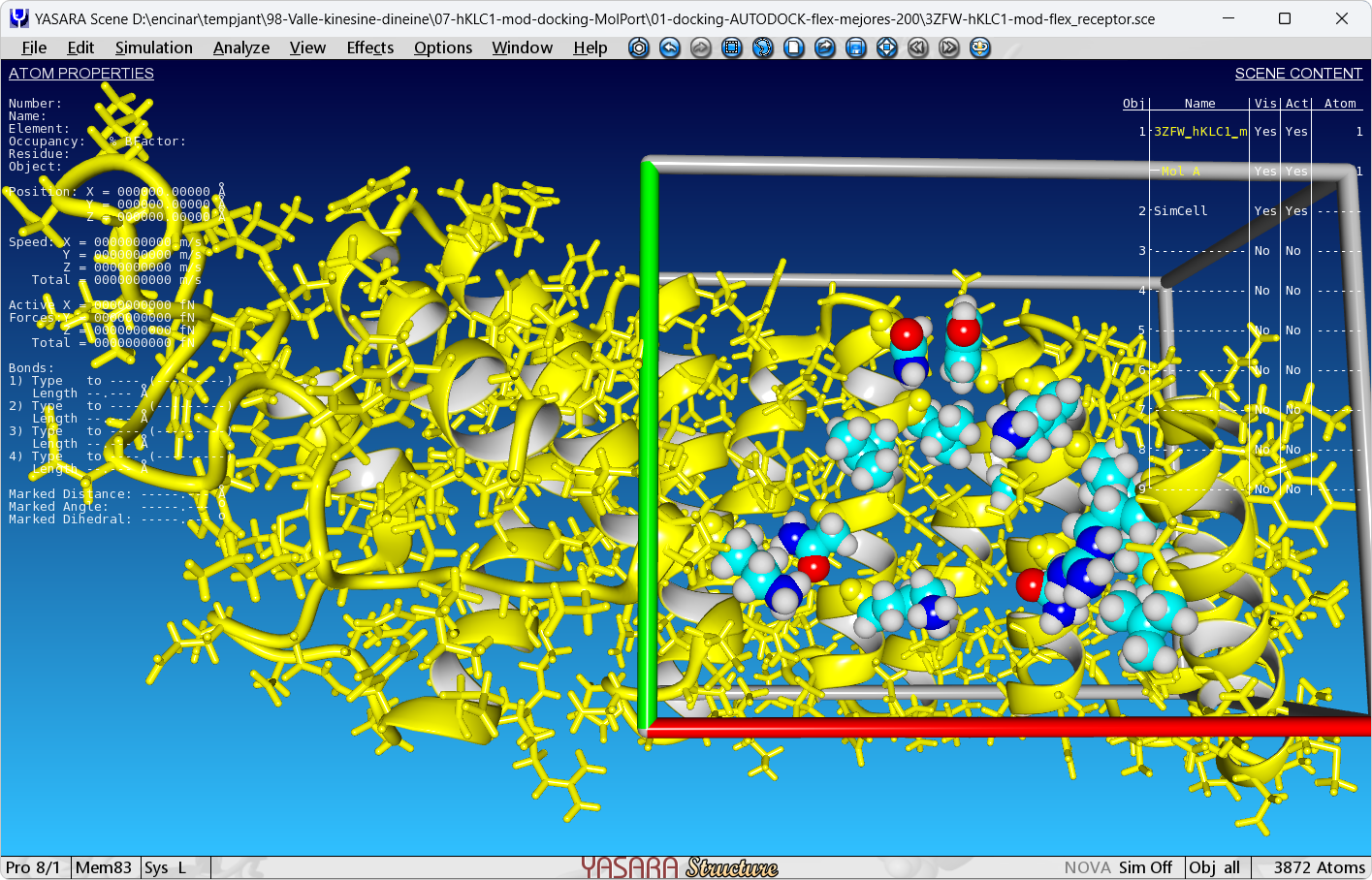
ForceField AMBER03Antes de ejecutar el macro dock_runscreening.mcr debemos preparamos una escena de YASARA (en este ejemplo 3ZFW-hKLC1-mod-flex_receptor.sce) fijando todos los átomos salvo los de las cadenas laterales del sitio de unión al péptido que tiene el cristal 3ZFW. Para ello, desde la consola de YASARA y después de cargar el fichero 3ZFW.pdb, ejecutamos las siguientes líneas:
FixRes Backbone
FixRes Sidechain
FreeAtom sidechain Res 278
FreeAtom sidechain Res 263
FreeAtom sidechain Res 266
FreeAtom sidechain Res 306
FreeAtom sidechain Res 302
FreeAtom sidechain Res 304
FreeAtom sidechain Res 309
FreeAtom sidechain Res 313
FreeAtom sidechain Res 340
FreeAtom sidechain Res 343
FreeAtom sidechain Res 347
FreeAtom sidechain Res 348
FreeAtom sidechain Res 351
FreeAtom sidechain Res 382
SelectRes Leu 278 Mol A or Leu 263 Mol A or Arg 266 Mol A or Leu 306 Mol A or Asn 302 Mol A or Ala 304 Mol A or Lys 309 Mol A or Tyr 313 Mol A or Lys 340 Mol A or Asn 343 Mol A or Leu 347 Mol A or Leu 348 Mol A or Asn 351 Mol A or Lys 382 Mol A
Cell Auto, Extension=3.0,Shape=Cuboid,selected
SaveSce 3ZFW-hKLC1-mod-flex_receptor.sce
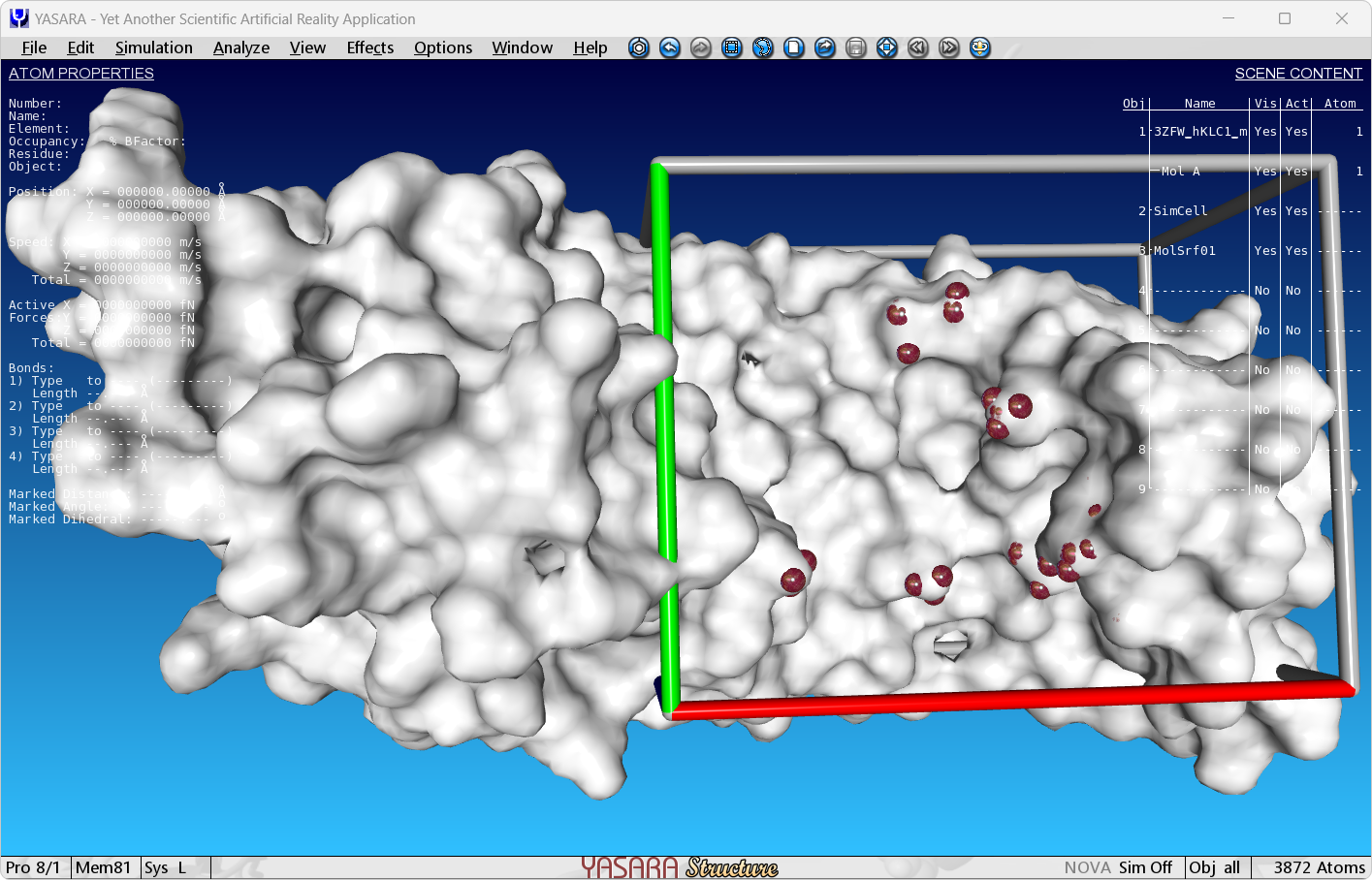
UnselectAll
BallAtom selected
ColorAtom selected,100
ShowSurfMol A or A,Type=Molecular,Update=Static,OutCol=FFFFFF,OutAlpha=100,Specular=Yes