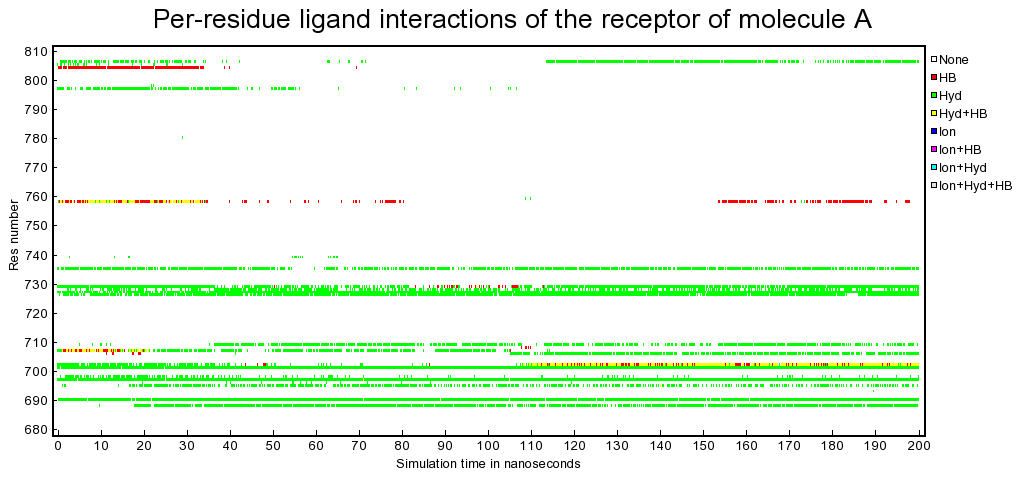
Fingerprint analysis of data from a protein-small compound molecular dynamics (MD) simulation.
matriz.txt
684 685 686 687 688 689 690 691 692 693 694 695 696 697 698 699 700 701 702 703 704 705 706 707 708 709 710 711 712 713 714 715 716 717 718 719 720 721 722 723 724 725 726 727 728 729 730 731 732 733 734 735 736 737 738 739 740 741 742 743 744 745 746 747 748 749 750 751 752 753 754 755 756 757 758 759 760 761 762 763 764 765 766 767 768 769 770 771 772 773 774 775 776 777 778 779 780 781 782 783 784 785 786 787 788 789 790 791 792 793 794 795 796 797 798 799 800 801 802 803 804 805 806 807
HB 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 9 0 0 0 1 3 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 3 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 23 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 17 0 0 0
Hyd 0 0 0 0 80 0 98 0 0 0 0 55 2 100 21 0 0 99 19 0 0 0 39 31 0 58 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 89 96 56 75 0 0 0 0 0 76 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 2 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 21 0 0 0 0 0 0 0 3 47 0
Hyd+HB 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 35 0 0 0 0 6 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 9 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Ion 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Ion+HB 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Ion+Hyd 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Ion+Hyd+HB 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0import pandas as pd
import matplotlib.pyplot as plt
# Entry route
entrada = r"d:\bbb\matriz.txt"
# We read the file with pandas
df = pd.read_csv(entrada, sep="\t")
# We extract interactions (first column, e.g. HB, Hyd, Ion...)
interacciones = df.iloc[:, 0]
# We extract amino acids (header except first column)
aminoacidos = df.columns[1:]
# We extract only numerical values
valores = df.iloc[:, 1:]
# Filter amino acids with interaction > 0
totales = valores.sum(axis=0)
aminoacidos_filtrados = [aa for aa in aminoacidos if totales[aa] > 0]
valores_filtrados = valores[aminoacidos_filtrados]
# Print to console
print("Tipo_interaccion\t" + "\t".join(aminoacidos_filtrados))
for idx, interaccion in enumerate(interacciones):
fila = [str(v) for v in valores_filtrados.iloc[idx].tolist()]
print(f"{interaccion}\t" + "\t".join(fila))
# Totals for filtered amino acids only
fila_total = [str(v) for v in valores_filtrados.sum(axis=0).tolist()]
print("TOTAL\t" + "\t".join(fila_total))
# Define custom colours
colores = {
"HB": "#FF0000",
"Hyd": "#00FF00",
"Hyd+HB": "#FFFF00",
"Ion": "#0000FF",
"Ion+HB": "#FF00FF",
"Ion+Hyd": "#00FFFF",
"Ion+Hyd+HB": "#E0E0E0"
}
# Assign colours in the same order as interactions
colores_lista = [colores.get(inter, "gray") for inter in interacciones]
# Stacked chart
ax = valores_filtrados.T.plot(
kind="bar",
stacked=True,
figsize=(12, 6),
color=colores_lista
)
# Change caption to show interaction names
ax.legend(interacciones, title="Interactions")
plt.xlabel("Aminoacids number")
plt.ylabel("Occupancy, %")
plt.title("Fingerprint of the small compound bound to the protein during MD simulation")
plt.xticks(rotation=90)
plt.tight_layout()
plt.show()
Interact 688 690 695 696 697 698 701 702 706 707 708 709 726 727 728 729 735 739 758 797 804 805 806
HB 0 0 0 0 0 0 0 9 1 3 1 0 0 0 0 3 0 0 23 0 17 0 0
Hyd 80 98 55 2 100 21 99 19 39 31 0 58 89 96 56 75 76 1 2 21 0 3 47
Hyd+HB 0 0 0 0 0 0 0 35 0 6 0 0 0 0 0 0 0 0 9 0 0 0 0
Ion 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Ion+HB 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Ion+Hyd 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
Ion+Hyd+HB 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
TOTAL 80 98 55 2 100 21 99 63 40 40 1 58 89 96 56 78 76 1 34 21 17 3 47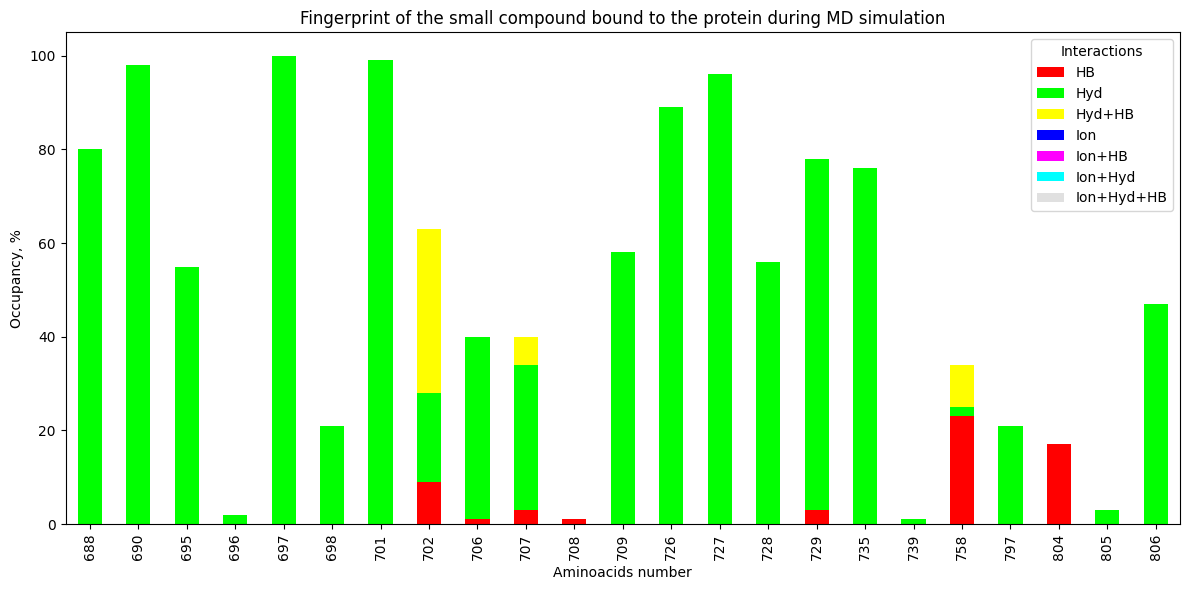
import re
import csv
from collections import defaultdict
def parse_file(path):
# Leer líneas (ignorando líneas vacías)
with open(path, 'r', encoding='utf-8-sig') as f:
raw_lines = [ln.rstrip('\n\r') for ln in f]
lines = [ln for ln in raw_lines if ln.strip() != '']
if not lines:
raise ValueError("El fichero está vacío o solo contiene líneas en blanco.")
# Heurística: si la primera línea contiene letras, la consideramos encabezado
header_line = lines[0]
has_header = bool(re.search(r'[A-Za-zÁÉÍÓÚáéíóúÑñ_\-]', header_line))
data_lines = lines[1:] if has_header else lines
# Tokenizar todas las líneas
tokenized = []
max_tokens = 0
for ln in data_lines:
toks = re.split(r'\s+', ln.strip())
tokenized.append(toks)
if len(toks) > max_tokens:
max_tokens = len(toks)
if max_tokens % 2 != 0:
max_tokens += 1
n_pairs = max_tokens // 2
if n_pairs == 0:
raise ValueError("No se han detectado pares residuo/valor en el fichero.")
# Nombres de condiciones
cond_names = []
if has_header:
h_tokens = re.split(r'\s+', header_line.strip())
if len(h_tokens) == 2 * n_pairs:
cond_names = [h_tokens[2*i + 1] for i in range(n_pairs)]
elif len(h_tokens) >= n_pairs:
cond_names = h_tokens[:n_pairs]
else:
cond_names = h_tokens + [f"Cond{i+1}" for i in range(len(h_tokens), n_pairs)]
else:
cond_names = [f"Cond{i+1}" for i in range(n_pairs)]
# Acumular datos
maps = [defaultdict(int) for _ in range(n_pairs)]
residues_set = set()
for toks in tokenized:
if len(toks) < 2 * n_pairs:
toks = toks + [''] * (2 * n_pairs - len(toks))
for i in range(n_pairs):
res_tok = toks[2*i].strip() if 2*i < len(toks) else ''
val_tok = toks[2*i + 1].strip() if 2*i + 1 < len(toks) else ''
if res_tok == '':
continue
try:
resid = int(float(res_tok))
except Exception:
continue
try:
val = int(float(val_tok)) if val_tok != '' else 0
except Exception:
val = 0
maps[i][resid] = val
residues_set.add(resid)
residues = sorted(residues_set)
rows = []
for r in residues:
row = [r] + [maps[i].get(r, 0) for i in range(n_pairs)]
rows.append(row)
return cond_names, rows
def write_out(path_out, cond_names, rows):
header = ['Residue'] + cond_names
with open(path_out, 'w', encoding='utf-8', newline='') as f:
writer = csv.writer(f, delimiter='\t')
writer.writerow(header)
for row in rows:
writer.writerow(row)
# --- CONFIGURACIÓN MANUAL ---
entrada = r"D:\aaa\matriz_vertical_todos_fingerprint.txt"
salida = r"D:\aaa\matriz_vertical_todos_fingerprint_unificada.txt"
# ----------------------------
cond_names, rows = parse_file(entrada)
write_out(salida, cond_names, rows)
print(f"Salida guardada en: {salida}")
print(f"Columnas detectadas: {['Residue'] + cond_names}")
print(f"Filas escritas: {len(rows)}")
D:\aaa\matriz_vertical_todos_fingerprint_unificada.txt
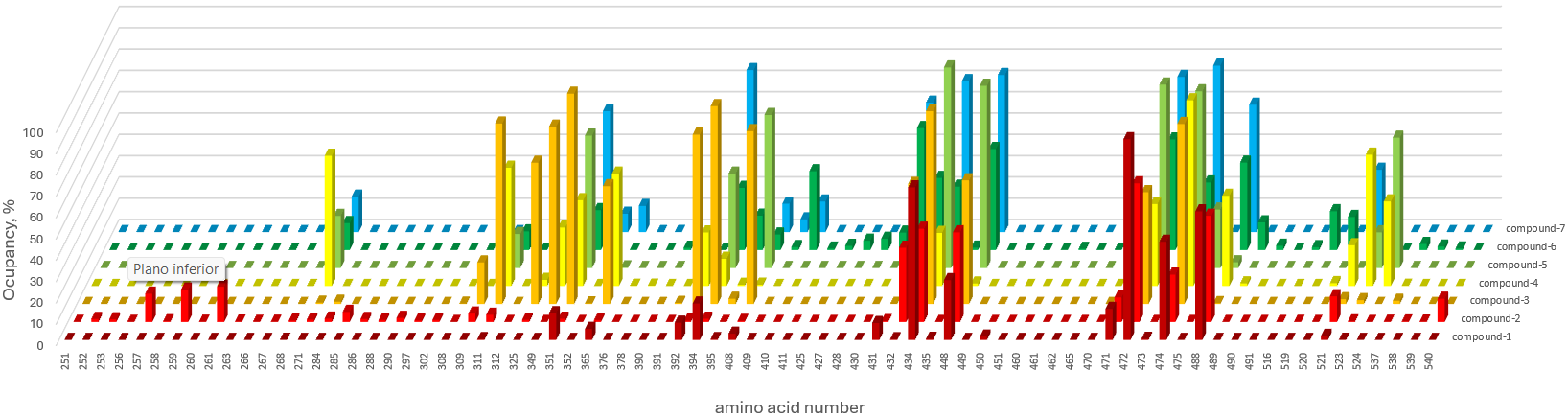
compound-1 compound-2 compound-3 compound-4 compound-5 compound-6 compound-7
251 0 0.05 0 0 0 0 0
252 0 1.4 0 0 0 0 0
253 0 1.25 0 0 0 0 0
256 0 0.05 0 0 0 0 0
257 0 13.29 0 0 0 0 0
258 0 0.35 0 0 0 0 0
259 0 15.19 0 0 0 0 0
260 0 0.1 0 0 0 0 0
261 0 16.54 0 0 0 0.05 0
263 0 0.2 0 0 0 0 0
266 0 0.05 0 0 0 0 0
267 0 0.1 0 0 0 0 0
268 0.05 0.7 0 0 0 3.8 0
271 0 1.05 0.6 61.12 24.49 12.64 16.64
284 0 1.6 1.1 0 0 0 0
285 0 4.75 0 0 0 0 0
286 0 1.4 0 0 0 0 0
288 0 0.95 0 0 0 0 0
290 0 1.9 0 0 0 0 0
297 0 0.75 0 0 0 0 0
302 0 0.95 0 0 0 0 0
308 0 0 0.15 0 0 0 0
309 0 3.95 19.49 0 0 0 0
311 0.4 3.4 84.71 55.52 15.99 8.8 6.8
312 0 0 0 0 0 0 0.3
325 0 1 66.27 2.9 0 0.1 0
349 0 0 83.21 27.44 0 0.2 0
351 12.54 1.5 98.7 40.28 62.12 18.74 56.72
352 0 0 0.3 0 0 4.55 8.6
365 5.15 0.9 55.27 52.72 0.3 0.15 12.29
376 0 0 0 0.1 0 0 0
378 0 0 0 0.15 0 0 0.05
390 0 0 0 0 0 1.05 8
391 0 0 0.5 0 53.02 0.25 3.7
392 8.1 0.65 79.46 25.14 37.38 0 11.69
394 17.09 1.55 92.85 12.79 44.33 29.19 76.01
395 0.4 0 2.5 0.55 2.25 16.14 20.14
408 2.95 0.5 81.06 0.6 71.81 7.3 13.34
409 0 0 0 0 0 1.4 5.95
410 0 0 0 0 0 36.98 14.34
411 0 0 0 0 0 0.7 0
425 0 0 0 0 0 2 0
427 0 0 0 0 0 4.4 0
428 0 0 0 0 0 5.3 0
430 0 0 0 0 0 8.2 0
431 7.95 0.4 0.05 0.05 29.84 57.27 60.72
432 0.35 34.83 56.32 0.4 38.68 33.83 29.84
434 71.56 43.73 90.45 24.94 94.15 29.74 70.86
435 0.5 0 0.1 0 10.59 1.15 0.05
448 28.04 42.08 58.02 1.2 85.51 47.43 73.71
449 0 0 0 0 0 0.05 0
450 1.6 0 0 0 0 0 0
451 0 0 0 0 0 0.1 0
460 0 0 0 0 0 0.05 0
461 0 0 0 0 0 0.55 0
462 0 0 0 0 0 0.1 0
465 0 0 0 0 0 0.05 0
470 0 0 0 0 0 12.44 0
471 14.54 11.54 0 3.75 0 14.94 0
472 94.2 65.07 52.47 38.48 86.11 51.92 72.81
473 0.5 0 0 0 0 0 0
474 46.08 22.24 84.46 87.16 82.96 31.73 78.11
475 0 0 0 0 27.44 0 0
488 60.32 49.83 0.4 42.33 2.7 41.08 59.77
489 0 0 0 1.25 0 12.99 0
490 0 0 0 0 0 2.05 0
491 0 0 0 0 0 0.05 0
516 0 0 0 0 0 1.35 0
519 0 0.05 0 0 0 18.24 0
520 0 0 0 1.35 0 15.54 0
521 1.9 12.04 2.25 19.14 0 29.69 29.24
523 0.05 0.3 1.8 61.62 16.74 19.59 29.14
524 0 0 0 39.68 61.17 0 0
537 0 0.1 1.5 0.1 0 2.9 0.1
538 0 0.25 0 0 0 1.65 0
539 0 0.2 0 0 0 0.75 0
540 0 10.89 0 0 0 0 0